

Journal of Medical Sciences and Health
DOI: 10.46347/jmsh.2019.v05i01.007
Year: 2019, Volume: 5, Issue: 1, Pages: 36-41
Original Article
Ashish Bajaj1, Bibhabati Mishra2, Poonam S Loomba2, Archana Thakur3, Abha Sharma4, Prachala G Rathod1, Madhusmita Das1, Ashna Bhasin1
1Senior Resident, Department of Microbiology, Govind Ballabh Pant Institute of Postgraduate Medical Education and Research, Delhi, India,
2Director Professor, Department of Microbiology, Govind Ballabh Pant Institute of Postgraduate Medical Education and Research, Delhi, India,
3Director Professor and HOD, Department of Microbiology, Govind Ballabh Pant Institute of Postgraduate Medical Education and Research, Delhi, India,
4Assistant Professor, Department of Microbiology, Govind Ballabh Pant Institute of Postgraduate Medical Education and Research, Delhi, India
Address for correspondence:
Dr. Ashish Bajaj, H. No. 364, Sector 31, Gurgaon - 122 001, Haryana, India. Phone: 09711459875. E-mail: [email protected]
Introduction: Increased mortality due to sepsis and bacteremia impacts health-care activities severely. Administration of broad-spectrum antibiotics empirically may lead to failure of treatment. Toxic effects of non-susceptible drugs can be harmful for the patients and lead to the development of resistance. Aim: The aim is to study the prevalence of multidrug-resistant Gram-negative bacteria causing bloodstream infections in a tertiary care center.
Materials and Methods: A total of 6265 blood samples were received in the Microbiology Department of GB Pant Hospital from January 1, 2018, to December 31, 2018. The samples were processed as per standard techniques. Identification and antimicrobial susceptibility testing were done by VITEK-2 Compact automated system and Kirby–Bauer disc diffusion method as per the Clinical and Laboratory Standards Institute guidelines.
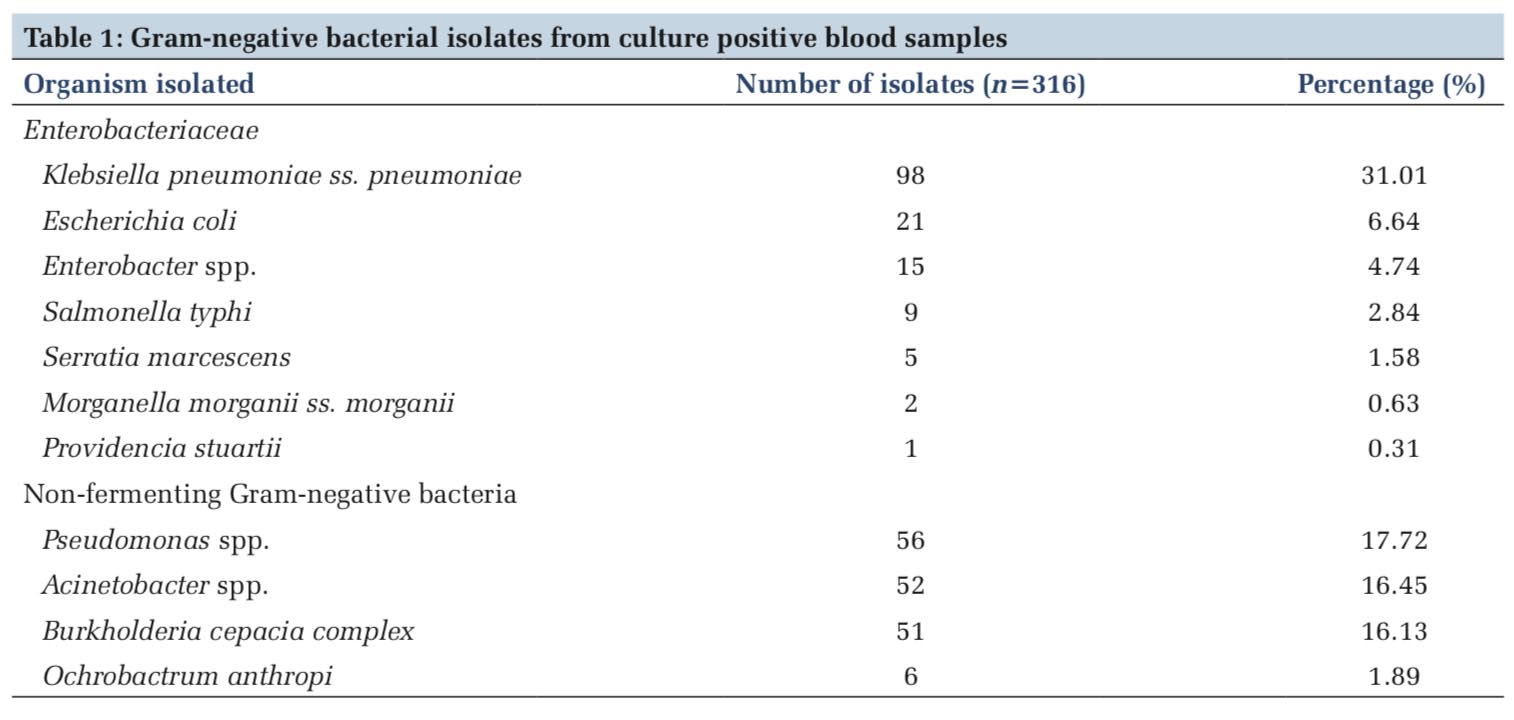
Results:Of total 6265 blood culture samples received in laboratory, 480 (7.66%) were culture positive. The Gram-negative bacteria 316 (65.83%) were isolated in majority followed by Gram-positive bacteria 148 (30.83%) and Candida spp. 16 (3.33%). Klebsiella pneumoniae (31.01%) was most common isolate among the Enterobacteriaceae. Whereas among the non-fermenting Gram-negative bacterial isolates, Pseudomonas spp. (17.72%) was most common. Gram-negative bacteria were resistant to commonly used antibiotics. 50–70% resistance was observed against carbapenems. Least resistance was seen to last resort antibiotics, i.e., tigecycline and colistin.
Conclusions:The increased isolation of multidrug-resistant Gram-negative bacteria is distressing, and further studies are advocated to help in the formulation of treatment and preventive strategies so as to curb such emergence.
KEY WORDS:Bloodstream infection, gram-negative bacteremia, sepsis.
Among the health care-associated infections, bacteremia accounts for maximum cases of mortality and morbidity.[1] Despite the vast improvement in diagnostic techniques, blood culture remains the gold standard for the diagnosis.[2]
The most frightening yet preventable complication in critical care units is bloodstream infection (BSI).
High incidence of multidrug-resistant bacteria leads to increased stay in hospital, rise in financial burden on the patient, and in many instances, loss of life.[3]
It is often associated with hospitalization, insertion of foreign bodies such as catheters into blood vessels, and other predisposing factors like stay in intensive care unit, lapses in hand washing, and non- adherence to infection control practices by medical staff. Genitourinary tract, intra-abdominal foci, and respiratory tract are the frequent sources of BSI.[4,5]
Blood culture bacterial isolates vary as per the geographical area. Changes in the local patterns of bacterial infection and susceptibility to various antibiotic should be critically evaluated periodically.[6]
The purpose of this study was to determine the antimicrobial susceptibility pattern of bacterial isolates from blood culture and determine the frequency of non-fermenters such as Pseudomonas aeruginosa and Enterobacteriaceae causing bloodstream infection. Increasing resistance to antibiotics and the emergence of multidrug-resistant organisms justify the need of this study. Similarly, as per local antibiogram, resistance to commonly used drugs such as cephalosporins, fluoroquinolones, aminoglycosides, and carbapenems was also increasing. Therefore, this study was aimed at finding the resistance pattern of the Gram-negative bacterial isolates from blood in a tertiary care center and formulate strategy for empirical treatment in septicemic patients.
Govind Ballabh Pant Institute of Postgraduate Medical Education and Research (GIPMER) is a superspecialty hospital providing care to patients from all over India. A total of 6265 blood samples were received in the Microbiology Department of GIPMER from January 1, 2018, to December 31, 2018.
Before administration of any antimicrobial therapy, blood culture sample was collected with aseptic precautions. 70% alcohol followed by 2% tincture iodine was used for surface disinfection at the site of collection. Adult and pediatric BACTEC blood culture bottles were inoculated with 10 ml and 3–5 ml of blood from adults and children, respectively. The bottles were then placed in BACTEC 9050 blood culture instrument (Becton Dickenson, USA) and incubated at 37°C. After BACTEC instrument flagged positive, the vials were subjected to Gram staining and further inoculation on sheep blood agar and MacConkey agar (HiMedia). The culture plates were then incubated aerobically at 37°C for 18–24 h. Identification and antimicrobial susceptibility testing were done by VITEK-2 compact automated system and Kirby–Bauer disc diffusion method on Mueller–Hinton agar as per the CLSI guidelines. Strains of Staphylococcus aureus (ATCC25923), Escherichia coli (ATCC25922), and P. aeruginosa (ATCC27853) were used for culture and susceptibility testing as controls.
Of total 6265 blood culture samples received in laboratory, 480 (7.66%) were culture positive. The Gram-negative bacteria 316 (65.83%) were isolated in majority followed by the Gram-positive organisms 148(30.83%). Among Gram-positive organisms, S. aureus (134) was most common isolate followed by Enterococcus spp. (14). Sixteen isolates (3.33%) of Candida spp. were also isolated. Among the Gram-negative bacterial isolates, 151 isolates were Enterobacteriaceae and 165 were non-fermenters. K. pneumoniae (98) was most common isolate among the Enterobacteriaceae followed by E. coli (21) and Enterobacter spp. (15). Among the non-fermenting Gram-negative bacteria, Pseudomonas spp. (56) was most common isolate followed by Burkholderia cepacia complex (52) and Acinetobacter spp. (51), respectively (Table 1).
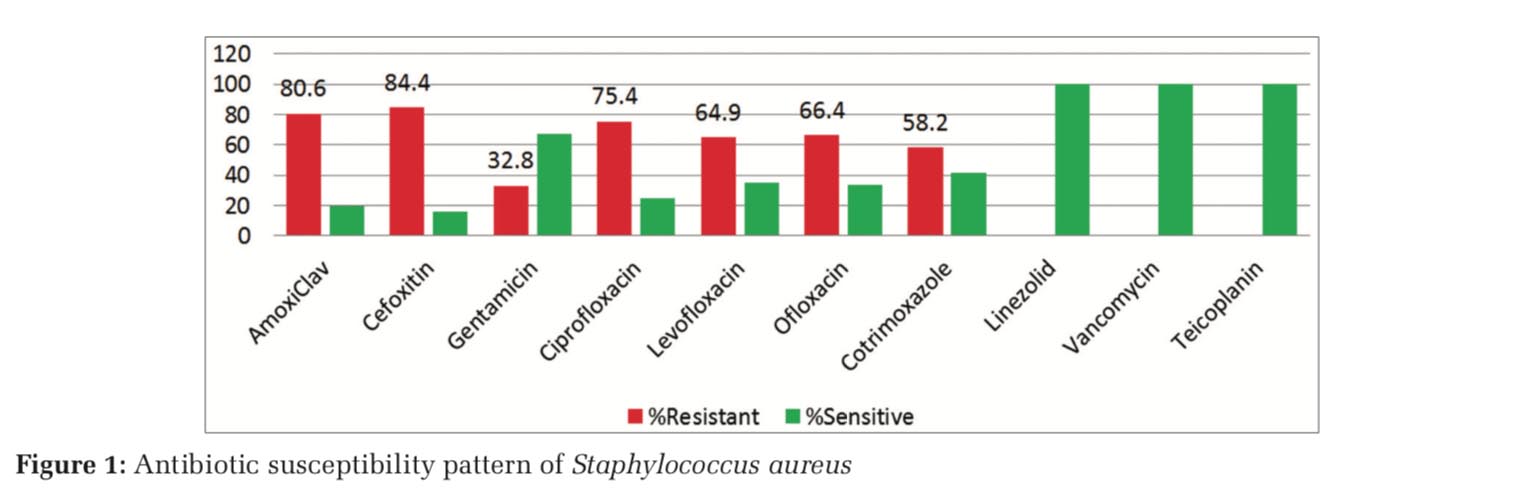
Among the Gram-positive isolates (148), S. aureus (134) was most common followed by Enterococcus spp. (14). The prevalence of methicillin-resistant S. aureus (MRSA) was 84.3%. Highest resistance was observed with amoxiclav followed by ciprofloxacin (Figure 1). Among the 14 isolates of Enterococcus spp., 92.8% were resistant to ampicillin and 71.4% to high-strength gentamicin. All Gram-positive bacterial isolates were sensitive to linezolid, vancomycin, and teicoplanin.
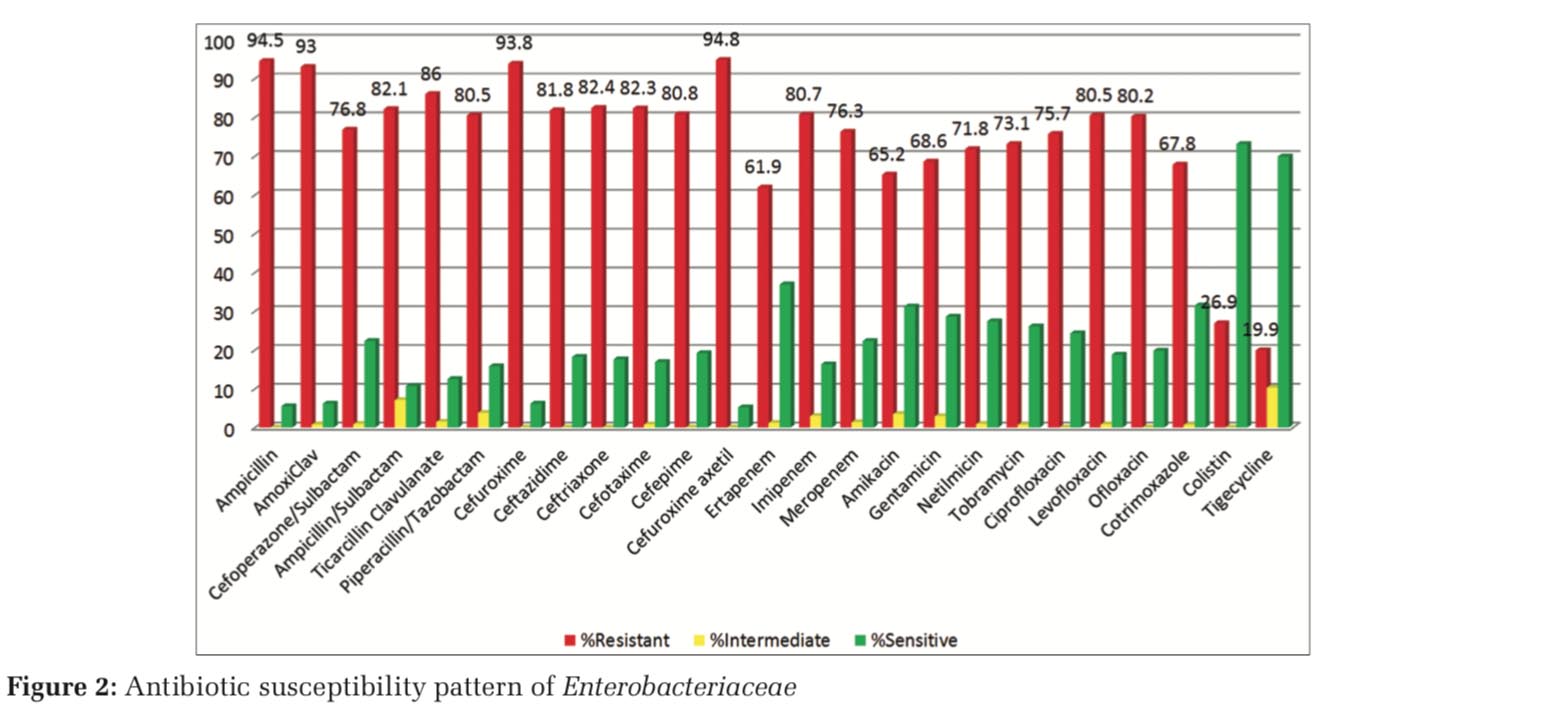
Gram-negative bacterial isolates were resistant to common antimicrobial agents. Among the Enterobacteriaceae family, 67%–80% resistance was observed for fluoroquinolones and more than 80% resistance for cephalosporins. Aminoglycosides showed around 65% resistance. 60%–80% resistance was observed against carbapenems (Figure 2).
Among the non-fermenting Gram-negative bacilli’s (NFGNBs), resistance varied from 50 to 80% for fluoroquinolones and from 80% to 90% for cephalosporins.Aminoglycosidesshowedaround60% resistance. 52–64% resistance was observed against carbapenems. Least resistance was seen to last resort antibiotics, i.e., tigecycline and colistin (Figure 3).
Burkholderia cepacia complex reported high resistance to ceftazidime, levofloxacin, and meropenem. Cotrimoxazole (83%) was found to be most sensitive drug (Figure 4).
Bloodstream infection is a life-threatening challenge; hence, timely detection, identification, and antimicrobial susceptibility testing of blood-borne pathogens are one of the most important functions of microbiology laboratory, especially in a tertiary care center.
GIPMER in New Delhi is a superspecialty hospital providing care to patients from all over India. The high prevalence of antimicrobial resistance rates in Delhi might be due to indiscriminate and over use of antibiotics in our country due to ease of their availability as highlighted by Roy et al. in his study.[7]
In this study, Gram-negative bacteria 316 (65.83%) were isolated in increased numbers as compared to the Gram-positive bacteria 148 (30.83%). Other studies also report Gram-negative bacteria as the most common cause of BSIs.[8,9]
Isolation of 16(3.33%) Candida spp. is similar to Kohli-Kochhar et al.[10] and Banik et al.[11] Among the Gram-positive bacterial isolates (148), S. aureus (134) was most common followed by Enterococcus spp. (14). Similar findings were reported by Banik et al.[11] The prevalence of MRSA was 84.3%. High susceptibility of Gram-positive bacterial isolates to vancomycin and teicoplanin is in conjunction with other studies.[11,12]
In the present study, K. pneumonia (98) was most common isolate among the Enterobacteriaceae family (151), followed by E. coli (21) and Enterobacter spp. (15). The antimicrobial susceptibility pattern, among the Enterobacteriaceae family, revealed a high level of resistance to common antimicrobials such as cephalosporins (80%) and fluoroquinolones (67–80%). Similar results were noted in study done by Swati et al.[13]
With regard to the third-generation cephalosporins, quinolones, beta-lactam, and beta-lactamase inhibitor combinations, the in vitro efficacy against members belonging to family Enterobacteriaceae did not reveal good results. These antibiotics have been used and abused to a significant extent in our health-care settings, thus making the base for the development of resistance. Gram-negative bacteria are being reported with significant increase in resistance to these group of antibiotics in studies done worldwide.[14-16]
Susceptibility (31.2%) to amikacin, however, revealed encouraging results against members of family Enterobacteriaceae similar to study done by Fayaaz et al., followed by tigecycline and colistin in this study.[17] 60–80% resistance was observed against carbapenems. Ertapenem was found to be most effective among carbapenems. With expeditious development of resistance to carbapenems by Enterobacteriaceae members, its use should be advocated only according to susceptibility report in the hospital.
Active surveillance is need of the hour in institutions treating immunocompromised patients to curtail the rapid emergence of antibiotic resistance and help formulate guidelines for empirical therapy. Monitoring of the development of antimicrobial resistance would positively boost ongoing regimens in developing country like India. Carbapenem susceptibility against all Gram-negative bacteria is less than reported in studies done in other part of the country.[18]
Among the NFGNBs (165), Pseudomonas spp. (56) was most common isolate followed by Burkholderia cepacia complex (52) and Acinetobacter spp. (51), respectively. Pseudomonas was reported as the most common isolate among the non-fermenters by Saghir et al.[19]
In case of NFGNB isolated from blood, netilmicin and tobramycin displayed better in vitro efficacy than carbapenems. Similarly, beta-lactam/beta- lactamase inhibitor combinations comprising ampicillin/sulbactam, piperacillin/tazobactam, and cefoperazone/sulbactam as well as carbapenems revealed better results when compared to cephalosporins and fluoroquinolones. These results are in conformity to work done at other centers.[14,15]
With rampant injudicious use of antibiotics and increase in carbapenem-resistant isolates, we need to look into strict compliance of antimicrobial stewardship program to avoid the catastrophe that can be caused by multidrug-resistant bugs. Active surveillance of resistance developing to antibiotics used in our hospital is the need of the hour for formulating better treatment strategy. Unless strict measures are implemented for promoting good prescription practices, the goal to control antibiotic resistance seems difficult. The emergence of multidrug-resistant Gram-negative organisms is alarming, and further studies are advocated to help in the formulation of treatment and preventive strategies so as to curb such emergence.
Subscribe now for latest articles and news.