Introduction
Type 2 diabetes mellitus (T2DM) is a metabolic disorder marked by hyperglycemia, insulin resistance, and relative insulin deficiency which results from the interaction between genetic and epigenetic factors 1. Cardiovascular disease (CVD) is a major cause of morbidity and mortality in T2DM patients 2. Various components like dyslipidemia, obesity, smoking, lack of exercise, alcohol intake, oxidative stress, and genetic factors have been reasoned as risk factors for CVD and T2DM 3. Oxidative stress is a basic leading factor for the occurrence of these cardiovascular complications 4. People with diabetes are 2 to 4 times more likely to develop coronary artery disease (CAD), 5 times more likely to develop peripheral vascular disease (PVD), and 2 to 6 times more likely to develop myocardial infarction (MI) or stroke than people without diabetes 2. Oxidative modification of low-density lipoprotein cholesterol (LDL-C) is adhered to be key for the pathogenesis of atherosclerosis 5 which consecutively results in coronary artery disease (CAD). Human paraoxonase-1(PON 1) is high-density lipoprotein cholesterol (HDL-C) associated enzyme that is soldered to the lipoprotein by its hydrophobic N terminal end and also restrained to Apo A-1 6, 7. The main source of PON1 is the liver and is seen associated with HDL-C in circulation accounting for its anti-oxidant effects to prevent oxidation of LDL-C 8. The activity of PON 1 is decreased in patients with T2DM as per previous studies 9, 10, 11, 12. Differences in PON 1 activity in the human population is due to the polymorphic distribution of an amino acid substitution in the active site of the enzyme 13, 14, 15. Additionally, the data from various studies showed ethnic variations in the interpretation of CVD and its association with PON 1 polymorphism 16, 17, 18. The human PON 1 gene is located on the long arm of chromosome 7 (7q21.3-q22.1) and human serum PON-1 (PON 1. EC 3.1.8.1) is a glycoprotein containing 355 amino acids 19, 20.
The two single nucleotide polymorphisms in the PON1 coding region that affect the anti-atherogenic properties and have been associated with risk for CVD. The PON1 gene polymorphism L55M (rs-854560) is linked to reduced activity of PON1, where leucine is substituted by methionine at amino acid position 55 of the third exon 21. This single nucleotide polymorphism in the coding region influences the anti-atherogenic property of PON1 22, 23. The other common polymorphism of the PON1 gene is Q192R (rs662) where glutamine (Q) is supplanted by arginine (R) in the coding region (exon 6) of the PON1 gene is substrate dependent and influences the hydrolytic activity of the serum PON1 to certain substrates including lipid peroxides 24, 25. Genetic studies exploring polymorphisms in critical genes such as PON1 have far-reaching implications. They help in understanding the molecular basis of diseases, such as how variations in gene sequences can predispose individuals to conditions like CVD in the context of T2DM. Such studies aid in identifying genetic markers that can predict disease risk, enabling more personalized approaches to prevention and treatment. Moreover, they pave the way for novel therapeutic interventions that target these genetic pathways. In the case of PON1, understanding its polymorphisms and resultant variations in enzyme activity may contribute to the development of antioxidant-based therapies aimed at mitigating cardiovascular complications. Since CVD complications are a major risk factor in diabetic patients, the identification of fundamental biochemical and molecular pathways that lead to cardiovascular complications and their association with paraoxonase-1 genes L55M and Q192R polymorphism in the Indian population with T2DM, along with their lipid profile, were explored in the present study.
The objectives of the study were set as (a) to determine the PON 1 gene frequency and its levels in the healthy controls. (b) to determine and compare the PON 1 gene frequency and its levels among newly diagnosed type 2 diabetic patients not on any hypoglycaemic treatment, those taking hypoglycaemic treatment for more than 6 months and T2DM patients with cardiovascular complications. (c) to determine the lipid profile of patients and controls.
Materials and Methods
Study population
This study was carried out in the Department of Biochemistry in association with the Department of Endocrinology, Vardhman Mahavir Medical College, and Safdarjung Hospital, New Delhi after getting approval from the Institutional Ethics Committee.
Using the results of study done by El-Lebedy et al. 26 the sample size is calculated as follows:
Sample size formula
4*( Zα + Zβ)2 / log (OR)
Zα = level of statistical significance (1.96) at CI = 95% Zβ = Desired power (0.84) at power of study =80%
OR = Odd’s ratio
By putting the values in the formula, the sample size comes out to be 56 and after adding 10% margin of error the minimum sample size comes out to be 67. But, in order to perform the polymorphism study, the sample size was taken as 100 as a sample size of around 100 is generally considered a reasonable starting point for most polymorphism studies. The study subjects were divided into four groups in the present study:
(1) T2DM subjects with fasting plasma glucose (FPG) ≥ 126 mg/dl, not on any hypoglycemic treatment.
(2) T2DM subjects with fasting plasma glucose (FPG) ≥ 126 mg/dl, on hypoglycemic treatment for more than 6 months duration.
(3) T2DM diagnosed subjects with FPG ≥ 126 mg/dl and had complications with any of the vascular diseases eg. Ischemic heart disease (IHD), macro-angiopathy and/or cerebrovascular disease
(4) Age and Sex matched controls with FPG < 100 mg/dl.
Sample collection
After informed written consent, a venous blood sample (6 ml) was collected from the study subjects after overnight fasting for further investigations. Total cholesterol (TC), Triglycerides (TG), High-density lipoprotein Cholesterol (HDL-C), Low-density lipoprotein Cholesterol (LDL-C), glucose along with LFT and KFT profiles were quantified using an Automated Clinical Chemistry Analyzer, Advia-2400 Siemens Pvt Limited (Germany). DNA was extracted manually from the nucleated blood cells with the help of the Krishgen DNA Extraction Kit. The DNA was then quantified by taking OD at 260nm on a spectrophotometer and the quality of DNA was checked in 1% agarose gel electrophoresis.
Estimation of PON1 levels
The serum samples from the study subjects were analyzed to estimate PON1 levels by ELISA method. Shanghai Coon Koon Biotech PON1 ELISA kit was used for the same. The ELISA kit was based on the principle of double antibody sandwich technology.
Genotyping
The specific single nucleotide polymorphism (SNP)s containing regions of the PON1 gene were amplified by PCR technique using their respective primers (Table 1). The amplification was done in a 20 μL PCR reaction mixture containing 25–30 ng genomic DNA, 50pmol of each primer, 10 μL DreamTaq green PCR Master-mix (Thermo-scientific) containing Dream Taq DNA polymerase, 2X DreamTaq Green buffer, 32 μM MgCl2, and 3.2 μM of each dNTPs (dATP, dCTP, dGTP and dTTP) using a thermal cycler (Himedia Eco-96). The PCR conditions used were as follows: initial denaturation at 95 °C for 5 min, followed by 30 cycles of denaturation at 95 °C for 30 sec; annealing at 56.2 °C (PON1 L55M), 60.7 °C (PON1 Q192R) for 30sec, then extension at 72 °C for 30sec, with a final extension step at 72 °C for 5 min. The respective amplified PCR products were incubated with Hin1II (for PON1 L55M), and MboI (for PON1 Q192R) restriction enzymes, and the digested products were then resolved on agarose gel electrophoresis (Table 2).
|
SNP
|
Primers
|
Product size
|
Genotyping
|
|
PON1 L55M (rs-854560) |
5’-AAGCCAGTCCATTAGGCAGT-3’(forward) |
135bp |
RFLP |
|
5’-CCCAGTTTCAAGTGAGGTGTG-3’(reverse) |
|||
|
PON1 Q192R (rs662) |
5′-GCT GTG GGA CCT GAG CAC TT-3′(forward) |
189bp |
RFLP |
|
5′-ATA CTT GCC ATC GGG TGA AATG-3’ (reverse) |
(RFLP: Restriction Fragment Length Polymorphism)
|
SNP
|
Restriction enzymes
|
Agarose gel electrophoresis
|
Fragment sizes
|
|
PON1 L55M (rs-854560) |
Hin1II (Thermo-scientific) |
1.2% |
LL- 135 bp |
|
LM- 135 bp, 106 bp, and 29 bp |
|||
|
MM -106 bp, 29bp |
|||
|
PON1 Q192R (rs662) |
MboI (Thermo-scientific) |
1.1% |
QQ- 156 bp and 33 bp |
|
QR- 189 bp,156 bp, and 33 bp |
|||
|
RR -189 bp |
Statistical Analysis
All statistical analyses were imposed using the Statistical Package for Social Sciences software (SPSS), version 25.0 (IBM, Chicago, USA) after the data was imported into a Microsoft Excel spreadsheet. The categorical variables were displayed as percentages (%) and numbers. Quantitative units were expressed as mean ± Standard Error of the Mean (SEM). One-way Analysis of variance (ANOVA) was applied to contrast the means of multiple independent study classes. Categorical variables were presented as percentages of respective study groups and rationalized with Pearson’s χ2 test to check the association between various genotypes of PON1 gene polymorphism and T2DM. The Odds Ratio (OR) was figured with a 95% confidence interval (CI). p-value < 0.05 was treated as statistically significant. All statistical analyses were imposed using the Statistical Package for Social Sciences software (SPSS), version 25.0 (IBM, Chicago, USA).
Results
Demographic Characteristics of research groups
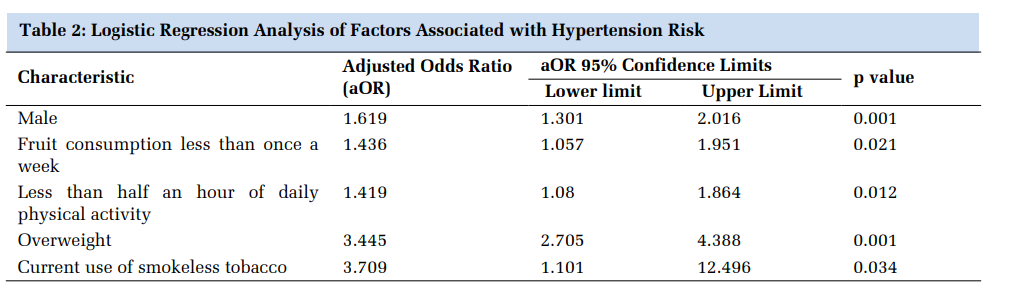
Table 3 depicts the demographic features and lipid profile in cases and controls.
The mean age of the newly diagnosed T2DM group not taking any hypoglycemic medication was 42.5, T2DM cases who were on oral hypoglycemic drugs for at least 6 months period was 48.57, T2DM with complications (CAD) group was 56.36, and controls group was 46 years. The percentage of males in the four groups was 28.57, 57.14, 57.14, 35.71 respectively. The case and the control groups (excluding T2DM with CAD group) and were age and gender-matched. There was no significant divergence delineated concerning triglycerides in the study groups. When total cholesterol was analyzed in the research groups, it was found to be significantly higher in the newly diagnosed T2DM group when collated with the control group (P value: <.001). In the case of HDL-C, there occurred a significantly lower level in T2DM with complications (CAD) group when compared with the controls (P value: .004). Strikingly, a similar pattern of significantly higher levels was etched concerning LDL-C (P value: <.001), TC: HDL ratio (P value: 0.04) in the newly diagnosed T2DM group, T2DM subjects on oral hypoglycemic drugs group while contrasting with the controls.
|
Attributes
|
Newly diagnosed T2DM group (n=14)
|
T2DM on oral hypoglycemic drugs for at least 6 months period group (n=14)
|
T2DM with complications (CAD) group (n=14)
|
Controls (n=14)
|
P value
|
|
Age (years±SEM) |
42.50 ± 1.98 |
48.57 ± 2.04 |
56.36 ± 2.30 |
46 ± 1.96 |
<.001 |
|
Age (years±SEM) |
42.50 ± 1.98 |
48.57 ± 2.04 |
|
46 ± 1.96 |
.11 |
|
Sex (%) |
|
|
|
|
|
|
Triglycerides (mg/dL ± SEM) |
141.36 ± 18.25 |
160.79 ± 19.07 |
122.14 ± 12.22 |
113 ± 10.79 |
.15 |
|
Total Cholesterol (mg/dL ± SEM) |
168 ± 9.24 |
143.93 ± 9.22 |
103.33 ± 7.75 |
124.49 ± 7.94 |
<.001 |
|
HDL-C (mg/dL ± SEM) |
41.71 ± 2.95 |
37.21 ± 2.08 |
29.66 ± 2.18 |
38.47 ± 1.79 |
.004 |
|
LDL-C (mg/dL ± SEM) |
127.79 ± 9.60 |
99.64 ± 10.43 |
59.31 ± 5.47 |
70.91 ± 4.61 |
<.001 |
|
TC:HDL ratio (ratio±SEM |
4.15 ± 0.23 |
3.91 ± 0.21 |
3.57 ± 0.26 |
3.26 ± 0.18 |
.04 |
Polymorphism analysis
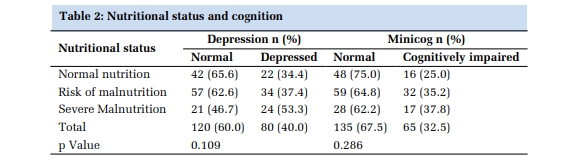
The extracted DNA was assessed for quality in 1% agarose gel electrophoresis (Figure 1). Following PCR amplification of PON1 Q192R (rs662), and PON1 L55M (rs854560) specific sites in DNA, the quality was scanned by resolving the PCR products in 1% agarose gel for the existence of 189bp, 135bp size sharp bands respectively (Figure 2, Figure 3). RFLP products were recognized in agarose gel electrophoresis for the presence of PON1 Q192R genotypes and PON1 L55M genotypes (Figure 4, Figure 5).





The distribution of genotypes and alleles for the PON1 rs662 (Q192R) polymorphism is shown in Table 4, Table 5 respectively. The frequencies of the AA, AG, and GG genotypes in T2DM subjects with complications group were 35.7%, 35.7%, and 28.6% respectively. The frequencies of the AA, AG, and GG genotypes in the control group were 42.9%, 50.0%, and 7.1% respectively. Further analysis showed that the homozygous GG genotype of PON1 rs662 SNP has 10.50 fold increased risk of developing T2DM with complications (CAD) in the Indian population (odds ratio OR = 10.50, 95% CI 0.91–121.39, p = 0.04). The wide confidence interval suggests high variability in the estimate, possibly due to a small sample size or rare events. The lower bound (0.91) is close to 1, meaning that while the OR suggests a strong association, the true effect might not be as strong or might even be null. A significantly higher frequency of the G allele was also observed in the T2DM subjects with complications group than in controls (OR 3.26, 95% CI 1.09–9.78, p = 0.03). The distribution of genotypes and alleles for the PON1 rs854560 (L55M) polymorphism is shown in Table 6, Table 7 respectively. No significant differences were prominent in the distribution of L55M genotypes among the controls and CAD patients.
|
Genotype frequency |
Newly diagnosed T2DM subjects, (n=14) |
T2DM (on oral hypoglycemic drugs for at least 6 months period), (n=14) |
T2DM subjects with complications (CAD), (n=14) |
All cases, (n=42) |
Controls, (n=14) |
P value |
|
AA |
5(35.7%) |
3(21.4%) |
4(28.6%) |
12(28.6%) |
6(42.9%) |
0.36 |
|
AG |
5(35.7%) |
8(57.1%) |
3(21.4%) |
16(38.1%) |
7(50.0%) |
|
|
Odds ratio (95% CI) |
0.86(0.16-4.47) |
2.29(0.41-12.73) |
0.64(0.10- 4.10) |
1.14(0.30- 4.29) |
|
|
|
GG |
4(28.6%) |
3(21.4%) |
7(50.0%) |
14(33.3%) |
1(7.1%) |
|
|
Odds ratio (95% CI) |
4.80(0.40- 58.02) |
6.00(0.42- 85.25) |
10.50(0.91- 121.39) |
7.00(0.74- 66.62) |
|
|
|
P value (cases vs controls) |
0.33 |
0.36 |
0.04 |
0.16 |
|
ODDs ratio, taking AA as reference; CI-confidence interval
|
Allelic frequency |
Newly diagnosed T2DM subjects, (n=14) |
T2DM (on oral hypoglycemic drugs for at least 6 months period), (n=14) |
T2DM subjects with complications (CAD), (n=14) |
All cases, (n=42) |
Controls, (n=14) |
value |
|
A |
15(53.6%) |
14(50.0%) |
11(39.3%) |
40(47.6%) |
19(67.9%) |
0.27 |
|
G |
13(46.4 %) |
14(50.0%) |
17(60.7%) |
44(52.4%) |
9(32.1%) |
|
|
Odds ratio (95% CI) |
1.83(0.62-5.42) |
2.11(0.71-6.25) |
3.26(1.09- 9.78) |
2.32(0.94- 5.72) |
|
|
|
P value (cases vs controls) |
0.27 |
0.17 |
0.03 |
0.06 |
|
|
Genotype frequency |
Newly diagnosed T2DM subjects, (n=14) |
T2DM (on oral hypoglycemic drugs for at least 6 months period), (n=14) |
T2DM subjects with complications (CAD), (n=14) |
All cases, (n=42) |
Controls, (n=14) |
P value |
|
AA |
7(50.0%) |
10(71.4%) |
5(35.7%) |
22(52.4%) |
9(64.3%) |
0.14 |
|
AT |
1(7.1%) |
2(14.3%) |
7 (50.0%) |
10(23.8%) |
3(21.4%) |
|
|
Odds ratio (95% CI) |
0.43(0.04-5.06) |
0.60(0.08-4.45) |
4.20(0.74-23.91) |
1.36(0.30-6.14) |
2(14.3%) |
|
|
TT |
6(42.9%) |
2(14.3%) |
2(14.3%) |
10(23.8%) |
|
|
|
Odds ratio (95% CI) |
3.86(0.59- 25.29) |
0.90(0.10-7.78) |
1.80(0.19-16.98) |
2.05(0.37-11.25) |
|
|
|
P value (cases vs controls) |
0.2 |
0.88 |
0.25 |
0.69 |
|
ODDs ratio, taking AA as reference; CI-confidence interval
|
Allelic frequency |
Newly diagnosed T2DM subjects, (n=14) |
T2DM (on oral hypoglycemic drugs for at least 6 months period), (n=14) |
T2DM subjects with complications (CAD), (n=14) |
All cases, (n=42) |
Controls, (n=14) |
P value |
|
A |
15(53.6%) |
22(78.6%) |
17(60.7%) |
54(64.3%) |
21(75.0%) |
0.25 |
|
T |
13(46.4%) |
6(21.4%) |
11(39.3%) |
30(35.7%) |
7(25.0%) |
|
|
Odds ratio (95% CI) |
2.60(0.84-8.07) |
0.82(0.24-2.84) |
1.94(0.62- 6.09) |
1.67(0.64- 4.37) |
|
|
|
P value (cases vs controls) |
0.09 |
0.75 |
0.25 |
0.3 |
|
|
In this study, while comparing the mean serum PON 1 levels in study groups, serum PON1 level in diabetic patients having complications (CAD) was significantly decreased compared to those without complications and healthy individuals (P <0.0001, Table 8). When we compared the serum PON 1 levels in different genotypes of PON1 (Q192R) gene polymorphism in different study groups, serum PON1 level in GG genotypes of diabetic patients having complications (CAD) was significantly decreased compared to other genotypes (P value: 0.0001, Table 9).
|
Parameter |
Newly diagnosed T2DM subjects , (n=14) |
T2DM (on oral hypoglycemic drugs for at least 6 months period) , (n=14) |
T2DM subjects with complications (CAD), (n=14) |
All cases, (n=42) |
Controls, (n=14) |
P value |
|
Mean serum PON 1 levels (mIU/ml) |
1102.71±175.41 |
1032.57±194.81 |
557.63±383.29 |
877.17±354.07 |
1244.56±241.76 |
<.0001 |
Data are presented as mean±sd. Serum levels of PON1 were determined by ELISA (mIU/mL). Comparison between the groupswas performed with one-way ANOVA.
|
Genotypes |
Serum PON1 levels (mIU/ml) |
|||
|
Controls (n=14) |
Newly diagnosed T2DM subjects , (n=14) |
T2DM (on oral hypoglycemic drugs for at least 6 months period) , (n=14) |
T2DM subjects with complications (CAD), (n=14) |
|
|
AA |
1376.9±249.7 |
1211.12± 188.77 |
1211.33± 342.0 |
1060.48± 183.27 |
|
AG |
1157.7±206.3 |
1036.81 ± 153.50 |
988.39± 136.6 |
518.47± 239.14 |
|
GG |
1058.455 |
1005.99± 92.77 |
971.6± 55.62 |
287.07± 166.46 |
|
P value |
0.19 |
0.126478 |
0.20 |
0.0001 |
Data are presented as mean±sd. Serum levels of PON1 were determined by ELISA (mIU/mL). Comparison between the groups was performed with one-way ANOVA.
Discussion
The pathophysiology of T2DM is influenced by a complex interplay between genetic and environmental factors, including diet, lifestyle, and exposure to stressors. 27
Recent advances in genetic research have unveiled the intricate role of genetic polymorphisms in modulating disease susceptibility and drug response in T2DM. Samajdar et al. 28 emphasized the critical role of pharmacogenomics in personalized medicine for diabetes management. The study highlighted how variations in genes encoding drug-metabolizing enzymes, drug transporters, and therapeutic targets influence the efficacy and safety of antidiabetic treatments. Such insights advocate for pre-emptive genotyping to guide clinical decision-making, optimize therapeutic efficacy, and reduce adverse drug reactions.
In addition to pharmacogenomics, studies have also highlighted the importance of genetic markers in predicting complications associated with T2DM, particularly cardiovascular diseases. The human paraoxonase-1 (PON1) gene, which plays a pivotal role in the antioxidant defense mechanism by preventing low-density lipoprotein (LDL) oxidation, has garnered significant research interest. Polymorphisms such as Q192R and L55M in the PON1 gene are linked to variations in enzyme activity and susceptibility to CVD in diabetic patients.
The high plasma levels of lipid peroxidation products in diabetic patients are attributed to higher susceptibility of lipoproteins for oxidation. Oxidative modification of LDL is believed to be a major triggering event in the initiation and progression of atherosclerosis and cardiovascular events 29. PON1 has received considerable attention in the last decade because of its potential role in antioxidation of LDL. Polymorphism in the PON1 gene may influence its gene expression and in turn its activity 30. In the present study, we investigated the association of PON1 Q192R and L55M polymorphisms along with the levels of serum PON1 levels in T2DM in Indian patients. Diabetic patients were divided into 3 groups: (i) Newly diagnosed T2DM patients, not on hypogycemic drugs T2DM; (ii) On oral hypoglycemic drugs for at least 6 months period (iii) T2DM with complications (CAD) group and compare them with non-diabetic Control subjects.
The present study revealed derangements in lipid profiles with respect to total cholesterol, HDL-C, LDL-C, TC: HDL ratio in cases than controls. These findings are in line with the earlier studies where deviations of lipid levels in T2DM diabetes was shown, despite good glycemic control 31, 32. The total cholesterol was found to be significantly higher in the newly diagnosed T2DM group not on hypoglycemic drugs, when collated with the control group in this study, which doesn’t occur with group 3 & group 4 of T2DM subjects, on oral hypoglycemic drugs for at least 6 months period group and T2DM with complications (CAD) group].
Treatment for dyslipidemia could be the reason for this finding in diabetic patients. Also, significantly lower level of HDL-C in T2DM with complications (CAD) group was seen as compared to the controls. This goes along with the previous multi-centre prospective study involving nearly 7000 patients 33 which concluded that low levels of HDL-C were associated with increased prevalence of DM and higher risk of CAD. We were also able to show that the homozygous GG genotype of PON1 rs662 SNP showed an increased risk of developing T2DM with complications (CAD), but this result was contradictory to the study done by Bhattacharyya et al, QQ192 genotype showed an increased cardiovascular risk when compared to study subjects with PON1 RR192 or QR192 genotype 34. However, Kotur et al study did not show any link between the Q192R polymorphism and CAD 35. A meta-analysis done by X Huo et al, concluded that the PON1 Q192R polymorphism is associated with a significantly increased risk of CAD in T2DM patients in both Asian and Caucasian populations 36. Our study did not support any association of PON1 L55M polymorphism with diabetes and CAD in line with the previous studies 37, 38, 39. PON1 protects from atherosclerosis by preventing LDL-C from oxidation and by hydrolyzing the oxidized LDL-C, proved by in-vitro and in-vivo studies 32. On similar lines, serum PON1 levels in diabetes with complications (CAD) group and GG genotypes of PON1 rs662 (Q192R) in the same group were significantly decreased in our study. PON-1 enzyme activity was also found to be lower in diabetic patients with complications when compared to those without complications 40.
The present study represents an elaborative report of PON1 Q192R, L55M polymorphisms and PON1 levels in association with T2DM and its complication, CAD. Lowered PON1 levels increases the risk of development of T2DM and its complications. Also, this study is the first step in understanding the complex interplay genetic factors in diabetic individuals categorised in various subgroups as newly diagnosed and not taking hypoglycemic drugs, patients on hypogycemic drugs for more than 6 months duration and diabetic individuals with complications (CAD). Furthermore, in the context of India, where genetic diversity is immense and the burden of T2DM and associated cardiovascular complications is rising, this research could be transformative. Understanding the specific PON1 gene polymorphisms prevalent in the Indian population may guide the development of targeted genetic tests to identify individuals at heightened risk for CVD. Early identification through genetic screening can enable timely lifestyle interventions and pharmacological treatments tailored to the genetic profile of patients. Additionally, personalized treatment strategies could be developed by incorporating genetic data to optimize the selection of hypoglycemic agents, antioxidant therapies, and lipid-lowering medications. This approach may significantly improve disease outcomes and reduce healthcare costs associated with managing long-term complications of diabetes.
However, the present study has limitations too. The study sample size could be one of them. The results are in line with some studies, but contradictory findings have also been observed with others. Further studies with larger sample sizes and more precise estimates are needed to confirm the association.
Conclusion
This study highlights the significant association between the PON1 Q192R polymorphism and the risk of cardiovascular disease in Type 2 Diabetes Mellitus (T2DM) patients within the Indian population. The findings indicate that the GG genotype of the Q192R polymorphism is associated with a heightened risk of coronary artery disease (CAD) in diabetic individuals, while the L55M polymorphism does not show a significant correlation. Furthermore, lower serum PON1 levels observed in T2DM patients with CAD suggest a potential role of oxidative stress in disease progression. These findings have important clinical implications. Genetic screening for PON1 polymorphisms could serve as a valuable tool for early identification of individuals at heightened risk of cardiovascular complications, allowing for targeted interventions and personalized management strategies. Moreover, therapeutic approaches aimed at enhancing PON1 activity, such as antioxidant-based therapies or lifestyle modifications, may hold promise in mitigating disease progression and improving patient outcomes. Given the growing burden of diabetes and its associated complications in the Indian population, further large-scale, multi-centered studies are warranted to validate these findings and explore their translational potential in precision medicine.
Disclosure
Funding
None
COnflict of Interest
None