

Journal of Medical Sciences and Health
DOI: 10.46347/jmsh.v10.i3.24.248
Year: 2024, Volume: 10, Issue: 3, Pages: 294-300
Original Article
Nivetha Subramanian1 , Mangaiyarkarasi Thiyagarajan2 , Udhayasankar Ranganathan3 , Sunil S Shivekar3 , R Gopal4
1Tutor, Department of Microbiology, Sri Manakula Vinayagar Medical College and Hospital, Kalitheerthalkuppam, Madagadipet, Pondicherry - 605107, India,
2Professor, Department of Microbiology, Sri Manakula Vinayagar Medical College and Hospital, Kalitheerthalkuppam, Madagadipet, Pondicherry - 605107, India,
3Associate Professor, Department of Microbiology, Sri Manakula Vinayagar Medical College and Hospital, Kalitheerthalkuppam, Madagadipet, Pondicherry - 605107, India,
4Professor and HOD, Department of microbiology, Sri Manakula Vinayagar Medical College and Hospital, Kalitheerthalkuppam, Madagadipet, Pondicherry - 605107, India
Address for correspondence:
Udhayasankar Ranganathan, Associate Professor, Department of Microbiology, Sri Manakula Vinayagar Medical College and Hospital, Kalitheerthalkuppam, Madagadipet, Pondicherry - 605107, India.
E-mail: [email protected]
Received Date:01 August 2024, Accepted Date:26 September 2024, Published Date:08 October 2024
Introduction: The treatment options for multi drug resistant (MDR) and extensively drug resistant (XDR) Gram negative bacterial infections are limited. Colistin based combination therapy remains the last option for such infections. The emerging resistance to colistin, especially the transferrable mcr-1 gene mediated resistance poses a global threat. Objectives: This study aims to determine the colistin susceptibility among the multi drug resistant (MDR) clinical isolates of Escherichia coli, Kebsiella pneumoniae and Pseudomonas aeruginosa and also to determine the prevalence of mcr-1 gene in the plasmids of colistin resistant isolates. Materials And Methods: This cross-sectional hospital-based study was conducted in 2021. Colistin broth disk elution and colistin agar dilution methods were used to determine the colistin susceptibility of MDR clinical isolates. Plasmid DNA was isolated from colistin-resistant isolates by alkaline lysis, and conventional PCR was performed to detect the mcr-1 gene. Statistical Analysis: Statistical analysis was performed using SPSS software version 2.7. The results are presented in tables and charts. Results: In total, 124 MDR isolates were included in this study. Among the 124 isolates, 12 (9.6%) were found to be resistant to colistin using phenotypic methods. PCR for plamid mediated mcr-1 gene was negative in all 12 MDR isolates. Conclusion: Colistin resistance is relatively less among the MDR clinical isolates in our study. Transferable colistin resistance due mcr-1 gene mutation is not detected among colistin resistant isolates. Hence, colistin can be used with caution adapting the national and international guidelines in situations where most of the other antibiotics are resistant.
KEY WORDS: Multi drug resistance, Colistin, Colistin broth disk elution, Colistin agar dilution, mcr-1 Gene.
The injudicious and widespread use of antimicrobial agents has led to the emergence of multi drug resistant (MDR) and extensively drug-resistant (XDR) bacterial isolates. The antibiotic options for treating such challenging infections are limited, including a few newer antimicrobial agents, and revived older antibiotics such as colistin and fosfomycin. The new-generation β-lactam/β-lactamase inhibitor 1 Colistin-based combinations remain the last resort for the treatment of multidrug-resistant gram-negative bacterial infections. However, intrinsic resistance to colistin in various organisms and the emergence of acquired resistance since its reintroduction limits its use. The acquired resistance to colistin can be chromosomal or plasmid-mediated; however, plasmid-mediated resistance is of concern as it can be easily transferred across bacterial species. In 2016, the first plasmid-mediated colistin resistance gene, mcr-1 (mobile colistin resistant), was detected in Escherichia coli and Klebsiella pneumoniae. Since then, several variants of mcr-1 and novel families of mcr genes have been identified in various samples. Ten plasmid-encoded mcr gene variations capable of horizontal transmission have been identified. 2 Only a few cases of plasmid-mediated colistin resistance have been reported in India. In this study, we aimed to determine colistin susceptibility among multidrug-resistant (MDR) clinical isolates of Escherichia coli, Klebsiella pneumoniae, and Pseudomonas aeruginosa, and to determine the prevalence of the mcr-1 gene in the plasmids of colistin-resistant isolates.
This cross-sectional hospital-based study was conducted over a period of 12 months from January to December 2021 in the Department of Microbiology of a rural tertiary health care center in Puducherry, India. Patients whose clinical samples yielded MDR Escherichia coli, Klebsiella pneumoniae, and Pseudomonas aeruginosa were included in the study. The sample size was 124, calculated using Epi info ver 2 software with the prevalence of colistin resistance in India as 8.8%. 3 The institute ethics committee approval (Study No.EC/32/2021) was obtained prior to commencement of the study.
Escherichia coli, Klebsiella pneumoniae and Pseudomonas aeruginosa were identified by conventional phenotypic methods. 4 AST was performed by Kirby Bauer disk diffusion method using CLSI 2021 guidelines. 5 The media and disks for the tests were procured from HiMedia Laboratories (Mumbai, India).
MDR was defined as nonsusceptibility to at least one agent in three or more antimicrobial categories as per standard international terminology created by European Centre for Disease Control (ECDC) and Centre for Disease Control and Prevention (CDC), Atlanta. 6
Colistin susceptibility testing was performed using two methods: the CLSI Colistin broth disk elution method and the Colistin agar dilution method. 5 The media, colistin disks, and colistin powder were procured from HiMedia Laboratories (Mumbai, India).
Three to five colonies of overnight culture on nutrient agar plates were added to 5 mL of sterile normal saline and incubated for 30 min at 37°C. The turbidity was adjusted to 0.5 McFarland standard. For each MDR isolate, four test tubes were taken and labelled as 1,2,3 and 4, to which 10 ml of cation-adjusted Mueller Hinton broth [CAMHB] was added. One colistin disk (10 µg) was added to the first tube, two disks to the second tube, and four disks to the third tube at final concentrations of 1 µg/ml, 2 µg/ml, and 4 µg/ml, respectively. The fourth tube acts as a growth control, to which no disk is added. All test tubes were vortexed for at least 30 min to elute colistin. To each tube, 50 µL of the standard inoculum was added, tightly capped, and vortexed gently. The standard inoculum was also inoculated into a blood agar plate for a purity check. The tubes and plates were then incubated overnight at 35 °C. MIC for each strain was calculated as the lowest concentration that completely inhibited bacterial growth (Figure 1).

Colistin agar plates (CAP) with 1µg/mL, 2µg/mL and 4µg/mL were prepared by adding 20, 40, and 80 µL colistin stock solution (7.5 mg colistin in 20 mL distilled water) to 20 ml of Muller Hinton Agar (MHA). Each CAP was divided into 10 parts and labelled. Ten microliters of the prepared inoculum were streaked on each 1µg/mL, 2µg/mL and 4µg/mL of labelled CAPs. The standard inoculum was also inoculated in blood agar plate for purity check. The plates were incubated at 35°C overnight for 16-20 hours. The colistin and purity plates were carefully examined with transmitted light for colony or light film growth, and the MIC was interpreted (Figure 2).

Escherichia coli NCTC 13846 was used for quality control in both methods.
Interpretation: Isolates with MIC ≤2 µg/ml were considered intermediate and isolates with MIC ≥4 µg/mL were interpreted as resistant to colistin according to CLSI 2021 guidelines. 5
Phenotypically, colistin-resistant strains were incubated overnight in LB (Luria-Bertani) broth and the alkaline lysis method was adapted for plasmid DNA extraction. 7 The final product was run on an agarose gel to detect the presence of plasmids. After confirming the presence of plasmids, the template DNA was amplified along with the NCTC Escherichia coli 13846 control to detect the presence of the mcr-1 gene. Conventional polymerase chain reaction (PCR) was performed by adapting the primers and cycling conditions used by Rebelo et al. 8 With a final PCR reaction volume of 25 µL, the mcr-1 gene was amplified using the primers, 5’-AGTCCGTTTGTTCTTGTGGC- 3’ as forward and 5’-AGATCCTTGGTCTCGGCTTG- 3’ as reverse and the PCR conditions, single cycle of initial denaturation at 94 °C for 15 min, 25 cycles of denaturation at 94 °C for 30 s, annealing at 58 °C for 90 s, extension at 72 °C for 60 s, and a single cycle of final extension at 72 °C for 10 min. The PCR products were subject to gel electrophoresis (1.5 % agarose gel and ethidium bromide 0.5 µg/mL) and the separated products were visualised under ultraviolet gel doc. The 320 bp products represented the mcr-1 gene.
The data were analyzed using the SPSS software version 2.7. The results have been presented as tables and figures. Chi-square test was used to test the association between the variables. A p value of < 0.05 was considered as statistically significant.
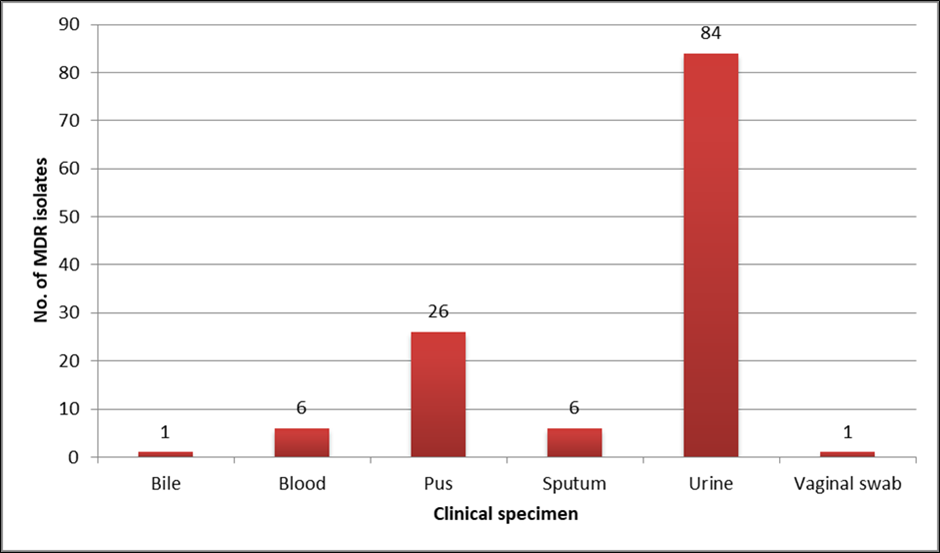
A total of 124 MDR isolates of Escherichia coli, Klebsiella pneumoniae and Pseudomonas aeruginosa were studied. There was no significant difference in distribution among gender, male 52% (n=65) and female 48%(n=59). Maximum isolates were among people aged more than 60 years (35.6%) (Table 1 ). Maximum isolates were from urine specimen (67.7%) (Figure 3 ). Escherichia coli, 29% (n=36) was the predominant MDR isolate. By colistin broth disk elution (CBDE) method, 12 isolates were resistant, and 112 were intermediate to colistin. Among the 112 intermediate isolates, two isolates had MIC 2 µg/ml and remaining 110 had MIC ≤ 1 µg/ml. By colistin agar dilution (CAD) method 12 were resistant and 112 were intermediate to colistin. All 112 isolates had MIC ≤ 1 µg/ml by CAD method. Colistin MIC of various MDR clinical isolates is given in Table 2. Conventional PCR for detection of mcr-1 plasmid gene was done for 12 isolates that were found to be resistant to colistin by both CBDE and CAD method. The two E. coli isolates with MIC 2 µg/ml by CBDE method was also included for PCR. All 14 isolates were found to be negative for mcr-1 gene.
|
Age Group in Years |
No. of Isolates (%) |
|
0-10 |
2 (1.6) |
|
11-20 |
5 (4) |
|
21-30 |
11 (8.8) |
|
31-40 |
19 (15.3) |
|
41-50 |
21 (17) |
|
51-60 |
22 (17.7) |
|
˃60 |
44 (35.6) |
|
Isolates |
Colistin Broth Disk Elution Method |
Colistin Agar Dilution Method |
||||
|
MIC |
MIC |
|||||
|
≤1 µg/mL
|
2 µg/mL
|
≥4 µg/mL
|
≤1 µg/mL
|
2 µg/mL
|
≥4 µg/mL
|
|
|
E. coli (n=74) |
61 (82.5) |
2 (2.7) |
11(14.8) |
63 (85.2) |
0 |
11 (14.8) |
|
K. pneumoniae (n=36) |
35 (97.3) |
0 |
1(2.7) |
35 (97.3) |
0 |
1(2.7) |
|
P. aeruginosa (n=14) |
14 (100) |
0 |
0 |
14 (100) |
0 |
0 |
|
TOTAL (n=124) |
110 (88.7) |
2 (1.6) |
12 (9.7) |
112 (90.3) |
0 |
12 (9.7) |
Plasma-mediated colistin resistance is a great threat across the globe because resistance plasmids can be easily transferred between bacteria. This horizontal transfer of colistin resistance genes, mainly mediated by the mcr group of genes, can lead to rapid rates of evolution of colistin-resistant strains, making colistin ineffective for the treatment of MDR and XDR gram-negative organisms. This threat became a reality when the mcr gene was first reported in China in E. coli isolated from pigs. 9 This study aimed to detect the mcr-1 gene in plasmids of phenotypically colistin-resistant MDR clinical isolates of E. coli, K. pneumoniae, and P. aeruginosa.
In our study, majority of MDR isolates were from urine samples followed by pus and blood. Many studies in literature show MDR isolates are common in urine samples. 10, 11 We have included urinary isolates for mcr-1 detection despite the fact that colistin is recommended only for extremely drug resistance organisms, where all preferred drugs are resistant with the aim of detecting tranferrable resistance alone. 12 Among the 124 MDR isolates in our study, the majority were Escherichia coli (59.6%). Many authors have also reported MDR Escherichia coli as the major pathogen in similar studies. 10, 13
In multiple investigations using different methodologies, the colistin resistance varied from 6% to 32%, with a growing trend of resistance seen in the last few years. 14, 15, 16, 17, 18, 19 The increased use of the antibiotic in recent times may be the cause of the developing trend in colistin resistance. Comparatively, our study's colistin resistance was shown to be 9.6%.
While some bacteria like Proteus species, Morganella morganii, Providencia species, Serratia marcescens, Chromobacterium species and Burkholderia cepacia are known to be intrinsically resistant to colistin, few Enterobacterales like E. coli, Klebsiella pneumoniae and non- fermenters like Pseudomonas aeruginosa and Acinetobacter species have developed acquired resistance to colistin. In our study, colistin resistance was commonly observed in Escherichia coli, followed by K. pneumoniae. No colistin resistance was observed in P. aeruginosa. Previous studies also show that acquired resistance to colistin is common in E. coli and K. pneumoniae and less in P. aeruginosa. 14, 17, 18, 20
Both CBDE method and CAD method detected 12 colistin resistant isolates. Among the remaining 112 intermediate isolates, CBDE identified two E. coli isolates that had higher MICs in addition, which were also later included in PCR. Even though the mcr-1 gene was absent in all the 14 isolates in our study, testing colistin susceptibility by a combination of methods would help in determining true resistance rates. Difference in the test performance characters of different phenotypic methods were also observed in previous studies, highlighting the importance of combination testing. 17, 21, 22
Subsequent to first isolation of plasmid mediated mcr-1 gene in animals from China, many studies from different countries across the world reported the transferrable mcr-1 genes in animals. 23, 24, 25, 26, 27 Many previous studies from India have also documented the presence of mcr genes in human clinical isolates. 28, 29, 30 Plamid borne mcr-1 gene was not detected by PCR among the phenotypically colistin resistance clinical isolates included in our study. Colistin resistance in these isolates can be attributed to other gene mutations which alters the lipid A of the organism like pmrA/pmrB, phoP/phoQ and mgrB, which is out of scope of the present study.
Colistin-based combination therapy is one of the recommended treatments for difficult-to-treat organisms such as carbapenemase-producing organisms. Hence, colistin should be used judiciously and should always be guided by antibiotic susceptibility testing when available.
Colistin resistance is relatively less among the MDR clinical isolates in our study. Transferable colistin resistance due to mcr-1 gene mutation is not detected among colistin resistant isolates. Hence, colistin can be used with caution adapting the national and international guidelines in situations where most of the other antibiotics are resistant.
The authors declare that there are no conflicts of interest.
This work received no specific grant from any funding agency.
The procedures followed in the study were in accordance with the ethical standards of the institutional ethics committee on human experimentation and with the Helsinki Declaration of 1975, as revised in 2000. The study was approved by the institute ethics committee (Study No.EC/32/2021)
All authors contributed to the following stages of research work.
Conception, design, data acquisition, data analysis and interpretation of data.
Drafting the article and revising the critical intellectual content.
Approval of the final version to be published.
Subscribe now for latest articles and news.