

Journal of Medical Sciences and Health
DOI: 10.46347/jmsh.v11.i2.24.240
Year: 2025, Volume: 11, Issue: 2, Pages: 203-209
Original Article
Laldinsangi1 , Anita Pandey2 , Peetam Singh3
1M.Sc. Scholar, Department of Microbiology, Subharti Medical College, Swami Vivekanand Subharti University, Meerut, 250005, Uttar Pradesh, India,
2Professor and Head, Department of Microbiology, Subharti Medical College, Swami Vivekanand Subharti University, Meerut, 250005, Uttar Pradesh, India,
3Assistant Professor, Department of Microbiology, Subharti Medical College, Swami Vivekanand Subharti University, Meerut, 250005, Uttar Pradesh, India
Address for correspondence:
Peetam Singh, Assistant Professor, Department of Microbiology, Subharti Medical College, Swami Vivekanand Subharti University, Meerut, 250005, Uttar Pradesh, India. E-mail: [email protected]
Received Date:28 July 2025, Accepted Date:01 March 2025, Published Date:05 September 2025
Background: The Non-fermenting Gram-negative bacilli are being increasingly reported. Few of them have emerged as a major cause of healthcare associated infections. They often pose a great therapeutic concern because of their multidrug resistant nature. The rare and emerging pathogens in this category which are intrinsically resistant to important antimicrobials need to be reliably identified to prevent therapy failure. This study was carried out to observe the species distribution profile and antimicrobial susceptibility pattern of these bacteria. Material and Methods: The clinical specimens collected from the patients were subjected to aerobic culture on appropriate culture media. The isolated colonies on culture media were presumptively identified by conventional methods. The species level identification and antimicrobial susceptibility testing was done by VITEK 2 compact automated system. Results: Among all non-fermenting Gram-negative bacilli, Acinetobacter baumannii was predominantly isolated comprising 42.22% followed by Pseudomonas aeruginosa (39.01%) and Burkholderia cepacia (5.80%). The majority of the isolates were isolated from endotracheal aspirate (26.79%), followed by pus (23.58%) and blood (17.53%). Acinetobacter species and Pseudomonas species were having good susceptibility against colistin. The rare and unusual pathogens like Stenotrophomonas maltophilia, Brevundimonas diminuta and Elizabethkingia meningoseptica were also reported highlighting the importance of automated methods of identification. Conclusion: The automated or molecular methods are important for reliable characterization of spectrum of pathogens as infrequent isolates are usually misidentified by conventional methods leading to treatment failure further added by intrinsic resistance among non-enterobacterales. The species distribution profile and susceptibility data can help clinicians to select appropriate antimicrobials for treatment.
Keywords
Non-enterobacterales, Non fermenting Gram negative bacilli, Healthcare-associated infections, VITEK 2
The non-enterobacterales bacteria are non-fermenting Gram-negative bacilli (NFGNB). They are taxonomically diverse group of aerobic and non–spore forming bacilli. They neither use carbohydrates as a source of energy nor degrade them through metabolic pathways other than fermentation. 1 They were considered to be contaminants in the past but have now emerged as significant healthcare-associated pathogens, especially in immunocompromised hosts. 2 Acinetobacter baumannii and Pseudomonas aeruginosa are the most commonly isolated NFGNB having a tendency to colonize various surfaces in healthcare settings which is crucial in their emergence as important nosocomial pathogens. 3 These pathogens may pose a serious problem by causing infections associated with indwelling medical devices like urinary catheter, humidifier and ventilator and have been implicated in a variety of life-threatening infections such as septicaemia, meningitis, bacteraemia, ventilator associated pneumonia (VAP), urinary tract infections (UTI), surgical site infections (SSI), wound infections, osteomyelitis, and meningitis. 4, 5 These non-enterobacterales bacteria exhibit resistant against many antimicrobials and have been documented to produce extended spectrum β-lactamases (ESBL) and metallo-β-lactamases (MBL). 6 The multidrug resistance (MDR) exhibited by these pathogens pose a major clinical problem in treating the infections caused by them. The conventional methods of identification are insufficient to identify these rare species. The misidentification is a common concern with conventional methods of identification leading to treatment failure due to intrinsic resistance exhibited by these bacterial species. The reliable species distribution trends of organisms and local antimicrobial susceptibility patterns of these emerging bacteria can help in selecting appropriate antimicrobial for effective empirical antimicrobial therapy and better outcomes. 7 This study may also provide a baseline information to formulate hospital antibiotic policy and also to prevent the spread of multidrug resistant strains within the hospital or in the community.
This cross-sectional hospital based observational study was conducted in the Department of Microbiology, Subharti Medical College, Swami Vivekanand Subharti University, Meerut for a period of one year, from May 2022 to April 2023. This study was approved by institutional ethics committee of Swami Vivekanand Subharti University (Medical), Meerut with reference number SMC/UECM/2022/400/202 dated 01/04/2022. The various clinical samples were collected by the trained personnel following standard precautions and aseptic procedures as per sample collection guidelines.
The clinical specimens collected from the patients were processed as per standard bacteriological techniques. 1 The clinical samples other than blood and urine were inoculated on blood agar, chocolate agar and MacConkey agar and incubated aerobically at 37°C for 24 to 48 hours. The urine samples were inoculated on cystine lactose electrolyte deficient (CLED) agar and incubated aerobically at 37°C for 24 hours. The blood culture was done by BacT/ ALERT 3D automated blood culture system (bioMerieux, France) using appropriate blood culture bottles for adults and paediatric age groups. The broth from positive flagged blood culture bottles was sub-cultured on blood agar, chocolate agar and MacConkey agar and incubated aerobically at 37°C for 24 to 48 hours. The isolates recovered from culture were provisionally identified by colony characteristic, morphology on Gram staining, motility, pigment production, catalase test and oxidase tests. The final species level identification and antimicrobial susceptibility testing (AST) was done by VITEK 2 compact automated system (bioMerieux, France) using GN test card and AST-N406 card respectively.
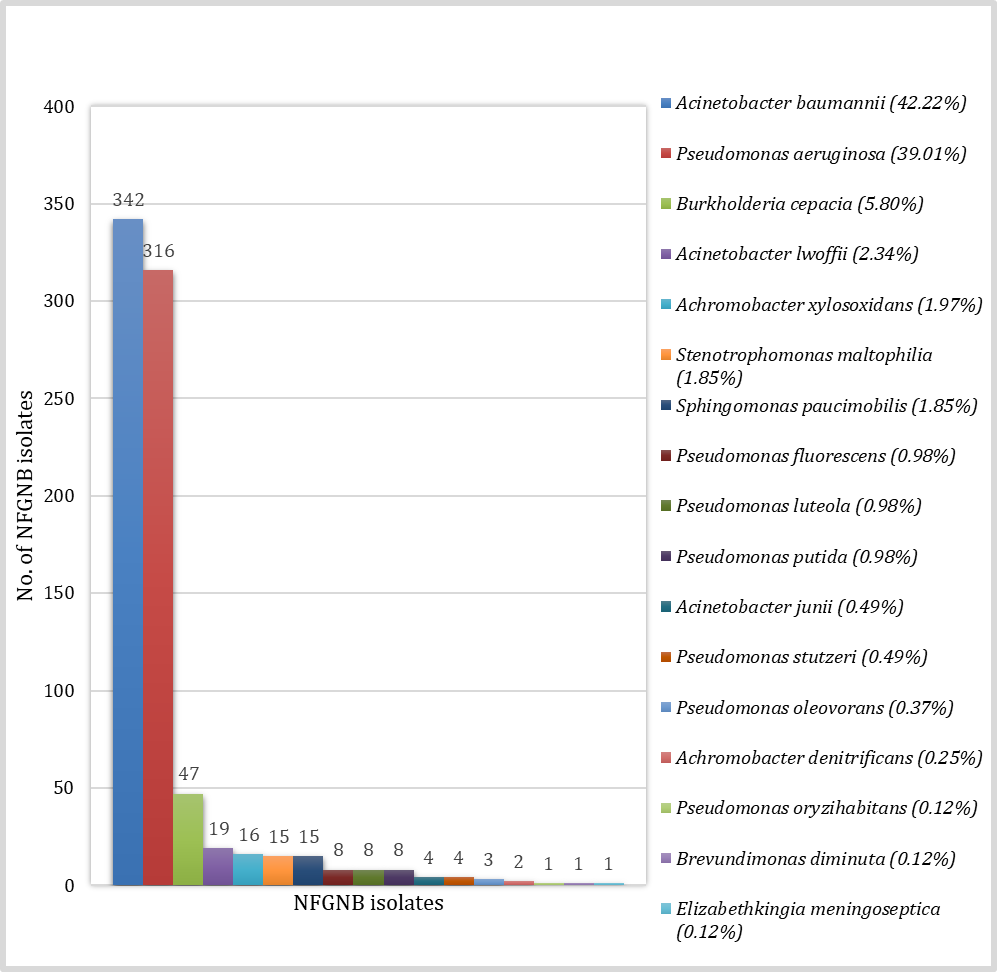
A total of 11897 samples were collected from the patients suspected of various clinical infections during the study period, out of which 5250 (44.12%) samples were culture positive. The bacterial isolates from all 5250 culture positive samples, 810 isolates were found NFGNB. The rate of isolation of NFGNB in our hospital was 15.43%. The NFGNB were isolated predominantly from indoor patients (73.08%) with male predominance (67.28%). The majority of the isolates were recovered from the patients belonging to the age group of 41-60 years (33.33%). The Acinetobacter baumannii was predominant bacterial isolate among NFGNB (42.22%) followed by Pseudomonas aeruginosa (39.01%). The species distribution of all NFGNB isolates is shown in Figure 1. The species level distribution of these NFGNB was not statistically significant at p-value of >0.05.
|
S. No. |
Organisms |
ET aspirate |
Pus |
Blood |
Sputum |
Urine |
BAL fluid |
Body fluids |
CSF |
Corneal scraping |
|
1. |
Acinetobacter baumannii (n= 342) |
137 |
54 |
65 |
32 |
21 |
20 |
9 |
4 |
0 |
|
2. |
Pseudomonas aeruginosa (n= 316) |
62 |
116 |
16 |
51 |
44 |
20 |
5 |
0 |
2 |
|
3. |
Burkholderia cepacia (n= 47) |
3 |
5 |
26 |
4 |
7 |
0 |
2 |
0 |
0 |
|
4. |
Acinetobacter lwoffii (n= 19) |
1 |
2 |
7 |
3 |
5 |
0 |
0 |
1 |
0 |
|
5. |
Achromobacter xylosoxidans (n= 16) |
2 |
0 |
12 |
2 |
0 |
0 |
0 |
0 |
0 |
|
6. |
Stenotrophomonas maltophilia (n= 15) |
5 |
6 |
2 |
0 |
0 |
2 |
0 |
0 |
0 |
|
7. |
Sphingomonas paucimobilis (n= 15) |
0 |
2 |
7 |
3 |
3 |
0 |
0 |
0 |
0 |
|
8. |
Pseudomonas fluorescens (n= 8) |
2 |
1 |
1 |
1 |
2 |
0 |
0 |
0 |
1 |
|
9. |
Pseudomonas luteola (n= 8) |
3 |
1 |
0 |
2 |
2 |
0 |
0 |
0 |
0 |
|
10. |
Pseudomonas putida (n= 8) |
1 |
3 |
1 |
0 |
1 |
2 |
0 |
0 |
0 |
|
11. |
Acinetobacter junii (n= 4) |
0 |
0 |
0 |
0 |
2 |
0 |
2 |
0 |
0 |
|
12. |
Pseudomonas stutzeri (n= 4) |
0 |
0 |
2 |
1 |
0 |
1 |
0 |
0 |
0 |
|
13. |
Pseudomonas oleovorans (n= 3) |
0 |
1 |
0 |
0 |
1 |
1 |
0 |
0 |
0 |
|
14. |
Achromobacter denitrificans (n= 2) |
0 |
0 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
|
15. |
Pseudomonas oryzihabitans (n= 1) |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
|
16. |
Brevundimonas diminuta (n= 1) |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
|
17. |
Elizabethkingia meningoseptica (n= 1) |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
|
No. of isolates |
217, 26.8 % |
191, 23.6 % |
142, 17.5% |
99, 12.2 % |
88, 10.9% |
47, 5.6% |
18, 2.2% |
5, 0.62% |
3, 0.37% |
|
|
Antibiot ics |
Acinetoba cter species (n= 365) |
Pseudom onas species (n= 348) |
Burkholde ria cepacia (n= 47) |
Achromoba cter species (n= 18) |
Stenotropho monas maltophilia (n= 15) |
Sphingomo nas paucimobilis (n= 15) |
Brevundim ons diminuta (n= 1) |
Elizabethkingia meningoseptica (n= 1) |
||||||||
|
R (%) |
S (%) |
R (%) |
S (%) |
R (%) |
S (%) |
R (%) |
S (%) |
R (%) |
S (%) |
R (%) |
S (%) |
R (%) |
S (%) |
R (%) |
S (%) |
|
|
PIT |
92 |
8 |
58 |
42 |
IR |
IR |
38 |
62 |
IR |
IR |
27 |
73 |
0 |
100 |
100 |
0 |
|
CIP |
91 |
9 |
71 |
29 |
- |
- |
66 |
34 |
- |
- |
47 |
53 |
0 |
100 |
100 |
0 |
|
TMP-SMX |
73 |
27 |
IR |
IR |
21 |
79 |
55 |
45 |
33 |
67 |
- |
- |
- |
- |
100 |
0 |
|
CAZ |
91 |
9 |
57 |
43 |
28 |
72 |
11 |
89 |
- |
- |
40 |
60 |
0 |
100 |
IR |
IR |
|
AT |
IR |
IR |
52 |
48 |
IR |
IR |
77 |
23 |
IR |
IR |
80 |
20 |
0 |
100 |
100 |
0 |
|
CPM |
84 |
16 |
47 |
53 |
IR |
IR |
17 |
83 |
- |
- |
13 |
87 |
0 |
100 |
100 |
0 |
|
GEN |
88 |
12 |
63 |
37 |
IR |
IR |
83 |
17 |
IR |
IR |
47 |
53 |
0 |
100 |
100 |
0 |
|
AK |
86 |
14 |
55 |
45 |
IR |
IR |
83 |
17 |
IR |
IR |
20 |
80 |
0 |
100 |
100 |
0 |
|
MI |
30 |
70 |
- |
- |
25 |
75 |
39 |
61 |
27 |
73 |
- |
- |
- |
- |
- |
- |
|
LE |
- |
- |
- |
- |
32 |
68 |
66 |
34 |
40 |
60 |
- |
- |
- |
- |
- |
- |
|
MRP |
90 |
10 |
57 |
43 |
15 |
85 |
22 |
78 |
IR |
IR |
27 |
73 |
0 |
100 |
IR |
IR |
|
IPM |
90 |
10 |
61 |
39 |
IR |
IR |
- |
- |
IR |
IR |
27 |
73 |
- |
- |
IR |
IR |
|
CL |
6 |
94 |
21 |
79 |
IR |
IR |
- |
- |
IR |
IR |
IR |
IR |
0 |
100 |
IR |
IR |
PIT= Piparacillin/ Tazobactum, CIP= Ciprofloxacin, TMP-SMX= Trimethoprim-sulfamethoxazole, CAZ= Ceftazidime, AT= Aztreonam, CPM= Cefepime, GEN= Gentamicin, AK= Amikacin, MI= Minocycline, LE= Levofloxacin, MRP= Meropenem, IPM= Imipenem, CL= Colistin, IR= Intrinsically resistant, S= Sensitive, R= Resistant
Overall, the NFGNB were isolated predominantly from the clinical samples of endotracheal aspirates (26.79%), followed by pus (23.58%) and blood (17.53%). The predominantly isolated NFGNB from endotracheal aspirate and pus were Acinetobacter baumannii and Pseudomonas aeruginosa respectively. The sample wise distribution of NFGNB is shown in Table 1. The sample wise distribution of NFGNB was not statistically significant at p-value of >0.05. The AST pattern of all clinical isolates of NFGNB against various antimicrobials is shown in Table 2. Overall, the Acinetobacter species exhibited higher resistance against carbapenems (90%) but good susceptibility against colistin (94%) and minocycline (70%). On the other hand, isolates of Pseudomonas species exhibited moderate susceptibility towards colistin (79%).
The rate of isolation of NFGNB in our study was found to be 15.43%. Slightly higher rate of isolation (18.8%) has been reported by Kumar et al. 8 However, Kishor et al, Kumar et al, Bansal et al and Soni et al reported lower isolation rate of 12.42%, 11.4%, 11.24% and 10.01% respectively. 9, 10, 11, 12 The frequency of isolation of various pathogens depends on multiple factors including the healthcare associated as well as patient related factors. We observed Acinetobacter baumannii (42.22%) as most common isolate among NFGNB followed by Pseudomonas aeruginosa (39.01%) and Burkholderia cepacia (5.80%). More or less similar findings were also reported by Kumar et al and Kishor et al. 8, 9, 11 The most of these studies did not report rare species including Achromobacter species, Sphingomonas paucimobilis, Brevundimonas diminuta, and Elizabethkingia meningoseptica. As these studies were based on the findings of conventional methods of identification, the probability of misidentification is high. The possibility of misidentification on conventional methods of identification is further supported by the studies based on automated identification including a study by Soni et al in which they also reported other rare pathogens. 12
In this study, majority of the NFGNB were isolated from endotracheal aspirate (ETA) comprising 26.8%, followed by pus (23.6%) and blood (17.5%). The Acinetobacter species were most commonly isolated from respiratory specimens (ETA) while Pseudomonas species were predominantly isolated from pus specimens. On the contrary, Soni et al and Gondha et al reported isolation of NFGNB most frequently from pus samples in their study. 12, 13 The majority of NFGNB in our study were isolated from samples received from admitted patients comprising 73.08% (42.22% from ICUs and 30.86% from wards) as compared to only 26.91% from OPD patients. The similar findings were also reported by Nazir et al. 14 The various risk factors included prolonged hospitalization in high-risk units, use of mechanical ventilation, underlying comorbidities such as burns, open wounds, surgical site infections, diabetes and malignancies highlighting the opportunistic nature of these pathogens.
The NFGNB were predominantly isolated from 40-60 years of age group, followed by young adults (21-40 years). The similar findings were also reported by Grewal et al and Berwal et al. 4, 15 On the contrary, Nazir et al reported majority of NFGNB in the age group of < 10 years. 14 There was male predominance (67.39%) in our study with male to female ratio of 2.06:1. The male predominance is in accordance with various other studies carried out by Gondha et al, Patel et al and Madkey et al. 13, 16, 17 However, Bansal et al and Berwal et al reported lower frequency of isolation among male patients as compared to female patients.11, 15 The variations in isolation rate of pathogens depending on demographic profile of patients may be due to variations in genetic and constitutional characteristics of the patients and associated comorbidities.
Most of the pathogens among NFGNB are intrinsically resistant to various antimicrobial groups and most of them are known to produce ESBLs and MBLs. 6 Most of the isolates in our study were MDR, posing a major clinical challenge in treating these. Acinetobacter baumannii exhibited higher resistance against carbapenems (90%) but good susceptibility against colistin (94%) and minocycline (70%). A comparable susceptibility to colistin and minocycline comprising 97.37% and 64.4% respectively was also reported by Soni et al. 12 Another study by Sengupta et al reported 100% susceptibility to colistin against Acinetobacter species.18 In this study, Pseudomonas species showed moderate susceptibility to colistin (79%) while only 39% isolates were showing susceptibility to carbapenems. On the contrary, Patel et al and Sarkar et al reported good susceptibility of Pseudomonas species to colistin, comprising 94.5% and 95% respectively. 16, 19
Contrary to our study, higher susceptibility of 70% to carbapenems was reported by Kumar et al, Gondha et al and Shah et al.10, 13, 20 The variations in resistance pattern of antimicrobials may vary from hospital to hospital as well as from community to community on one hand while variations due to falsely reported results as a consequence of lack of recommended methods of testing especially against colistin.
The clinical isolates of Burkholderia cepacia complex exhibited good sensitivity towards meropenem (85%), trimethoprim-sulfamethoxazole (79%), minocycline (75%) and ceftazidime (72%). Almost similar findings were also reported in a study by Nazir et al in which they reported a good susceptibility of Burkholderia cepacia against meropenem and minocycline comprising 67.7% and 70% respectively. 14 While another study by Shah et al reported all the isolates of Burkholderia cepacia as sensitive against meropenem. 20 Our clinical isolates of Stenotrophomonas maltophilia showed moderate susceptibility to minocycline (73%), trimethoprim-sulfamethoxazole (67%) and levofloxacin (60%). However, Sannathimmappa et al reported much higher susceptibility of Stenotrophomonas maltophilia to minocycline (97%), trimethoprim-sulfamethoxazole (93%) and levofloxacin (92%). 21 Though the three drugs are promising therapeutic options for treatment of infections caused by Stenotrophomonas maltophilia, the intrinsic resistance of Burkholderia cepacia and Stenotrophomonas maltophilia towards majority of the drugs contribute to their ability to survive in diverse environments and evade antimicrobial therapy leading to frequent treatment failure.
The other less commonly isolated NFGNBs such as Achromobacter species and Sphingomonas paucimobilis showed varied susceptibility to different groups of antibiotics as shown in Table 2. More or less similar findings were also reported previously in different studies. 22, 23, 24 The Achromobacter species were predominantly isolated from blood samples comprising 87.5 % isolates of Achromobacter species. Almost similar findings were reported recently by Siddique et al and Kar et al in 2023. 23, 24 Our data on sensitivity for Sphingomonas paucimobilis were similar to that reported by Rohilla et al where most of the isolates were having good susceptibility to cefepime. 25
Our isolate of Brevundimonas diminuta recovered from blood culture showed 100% sensitivity towards all the drugs, the resistance pattern was concordant with the study by Rani et al from the similar geographical region. 26 However, another single isolate of Elizabethkingia meningoseptica isolated from ETA showed pan-resistance against all the antimicrobials tested. In contrast to our study, Singh et al and Ganesan et al reported their Elizabethkingia meningoseptica exhibiting 100% susceptibility against minocycline and varied susceptibility against other drugs. 27, 28 This organism though isolated less frequently is known to survive in chlorine treated municipal water supplies, so it often colonizes sink basins and taps inside hospital settings and thus is a significant nosocomial threat.
The rare and emerging isolates reported in our study included Burkholderia cepacia, Stenotrophomonas maltophilia, Sphingomonas paucimobilis, and Elizabethkingia meningoseptica and all of these are intrinsically resistance to colistin. The misidentification will result in treatment failure if colistin is wrongly selected due to falsely reported colistin susceptibility. The possibility of misidentification of rare and intrinsically resistant NFGNB isolates on conventional testing is supported by many studies employing automated methods where these isolates were reported as compared to the studies employing conventional methods of identification usually missing out these isolates. 8, 9, 11, 12, 27, 28 We found that the changing trends as observed in this study highlighting the emergence of rare and intrinsically resistant isolates of NFGNB are in favour of exploration of the hidden instead of the changing trends.
This study was associated with certain limitations including the molecular characterization of species level identification of NFGNB isolates as well as genetic basis of resistance was not studied due to limited resources.
The automated or molecular methods are important for reliable identification and characterization of spectrum of pathogens as infrequent isolates are usually misidentified by conventional methods. The misidentification of NFGNB, especially those exhibiting intrinsic resistance to commonly used antimicrobials result in treatment failure due to wrongly selected intrinsically resistant antimicrobials. The reliable identification and susceptibility data can help clinicians while selecting appropriate antimicrobial treatment. The multidrug resistance exhibited by NFGNB pose great therapeutic concern as they have potential to survive in hospital environment leading to significant nosocomial threat. Therefore, identification and regular antimicrobial susceptibility surveillance of NFGNB, improved antibiotic stewardship and infection control measures are the need of the hour to control the emergence and spread of MDR strains of NFGNB in health care settings.
Nil.
None declared.
Subscribe now for latest articles and news.